Event
Our international symposium will be scheduled for March 14-16, 2026, followed by related two international conferences at the same venue:
- March 14-16: EPIC Assembly 2026 Symposium
- March 17: JST CREST Symposium
- March 18-19: Life-Nonlife Transition / Universal Biology Institute 2026 Symposium
- Event 2026.03: We will hold an international symposium. We warmly welcome your participation. → [Upcoming Event]
- Paper 2025.10: A paper by Group B02 (Mathematical Informatics) was published in Physical Review E. It proposes a method for graph coarse‑graining of chemical reaction hypergraphs based on conserved quantities and discusses the relationships of associated thermodynamic variables. The approach is expected to facilitate efficient analysis of diverse reaction assemblies. → [LINK]
- Paper 2025.09: A study by Group A02 establishing a quantitative method for dissociation of Cyclin–CDK complexes was published in Journal of Cell Science, advancing quantitative understanding of cell‑cycle control. → [LINK]
- Paper 2025.09: A theoretical model study by Group B01 on proliferating self‑propelled particle systems appeared in Physical Review Research, revealing a proliferation‑driven mechanism for the emergence of ordered motion without alignment interactions. → [LINK]
- Paper 2025.09: A theoretical proposal by Group B01 showing that light irradiation can induce nonreciprocal spin interactions in solids was published in Nature Communications, suggesting that the physics of nonreciprocal interactions observed in biological and ecological systems may be realized in solids. → [Original] → [Commentary]
- Paper 2025.09: Group B02 (Mathematical Informatics) published a general control theory in PRX Life for deriving optimal control laws for diverse biological populations, offering a methodology to analyze functional acquisition by biological assemblies from an optimization viewpoint. → [LINK]
- Event 2025.09: At IIBMP2025, we will co‑host Workshop WS‑8 “New Developments in Bioinformatics Inspired by Physics and Evolution,” September 4 (Thu) 15:40–17:10, Room 2. Results from evolution, information, and physics in our Area will be presented. We warmly welcome your participation! → [LINK]
- Info 2025.07: The application guidelines for publicly offered (open call) groups in Transformative Research Area A have been released. The description of our Area appears on p.55 of the official document. Please also see the explanatory article below. [Zenn article] → [LINK]
- Event 2025.07: We will hold an online information session on the publicly offered groups on July 22 (Tue) via Zoom. Please register from the Area website. → [LINK]
- Info 2025.06: An article introducing the aims and background of the Area has been published on Zenn, presenting our vision together with related references. → [LINK]
- Event 2025.09: The Area will co‑host a workshop “New Developments in Bioinformatics Inspired by Physics and Evolution” at the 2025 Annual Meeting of the Japanese Society for Bioinformatics (JSBi), September 3–5, Nagoya University. → [LINK]
- Event 2025.06: June 28–29: Toyota Konpon‑Ken Exploration Project “Principles and Limits of Unexplored Exploration” Open Symposium. → [LINK]
- Event 2025.06: June 28 (morning): “Evolutionary Information Assembly” Area Introduction Symposium. → [LINK]
- Paper 2025.04: An article by core members 澤井・梅津 was published in the April issue of Experimental Medicine (in Japanese), covering topics closely related to our Area. → [LINK]
- Info 2025.04: We launched the Area website.
- Info 2025.04: Transformative Research Area A “Principles of the Emergence of Biological Functions through Evolutionary Information Assembly (EPIC Assembly)” has started. Our target is to elucidate the principles by which advanced biological functions emerge and become refined.
Overview of the Research Area
Outline
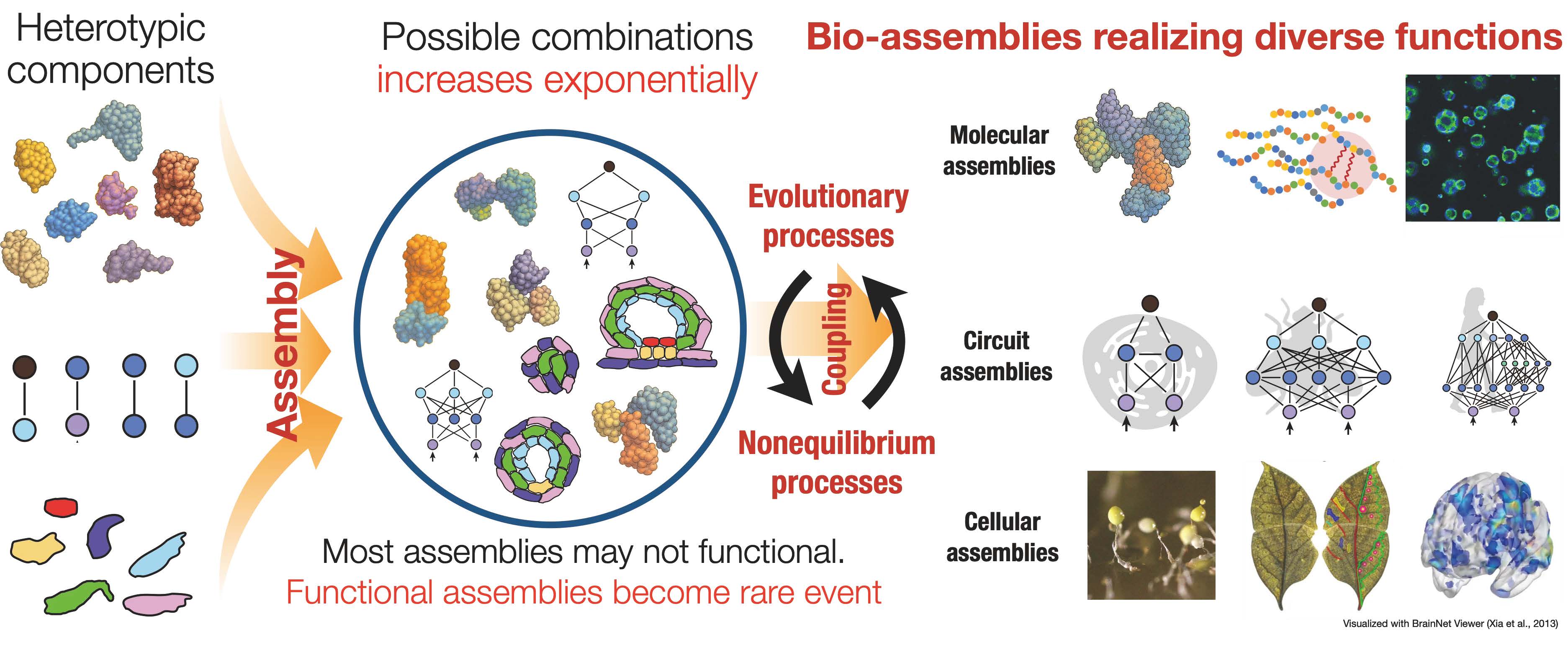
Biological systems exhibit a wide range of highly functional features. This research area aims to elucidate the principles underlying the emergence and development of such biological functions through evolution. To address the increase in complexity and diversity of the functions, we focus on the concept of "assemblies" as a unifying framework that spans molecular to multicellular levels. Assemblies refer to systems composed of heterotypic interacting elements. Assemblies can become combinatorially complex as the number and diversity of constituent elements increase, offering a structural basis for the evolutionary exploration of novel functions.
However, this combinatorial potential also poses a fundamental challenge: as the number of elements increases, the number of possible assemblies grows exponentially. How biological systems navigate this immense space to arrive at functional configurations remains an open question. We hypothesize that functional assemblies emerge and evolve through non-equilibrium dynamics, and that such dynamics are coordinated with evolutionary processes. Our goal is to understand the conjugate actions of non-equilibrium and evolutionary dynamics that drive the emergence, development, and increasing complexity of novel functions.
Research Strategy
To address this challenge, we integrate state-of-the-art imaging technologies and evolutionary analysis. Recent advances have enabled high-resolution measurement of dynamics across scales. In addition, next-generation sequencing and novel phylogenetic techniques allow the reconstruction of ancestral assemblies, whose functions will be verified by their experimental reconstitution.
We also construct mathematical frameworks to capture both the non-equilibrium and evolutionary dynamics of diverse assemblies, integrate these models with the experimental data, and identify the general principles by which non-equilibrium and evolution contribute to the emergence of novel functions in biology.
Expected Research Achievements
The experimental, computational, and theoretical methodologies developed in this area are expected to contribute to the design of various biological systems, such as synthetic smart droplets, information-processing biochemical circuits, and functional organoids. Furthermore, the theoretical insights into the interface of information, evolution, and non-equilibrium dynamics may open up novel theoretical paradigms for intelligent systems in information sciences.
Planned Members
Publicly Offered Members
Publications from this Area
Events Organized in this Area

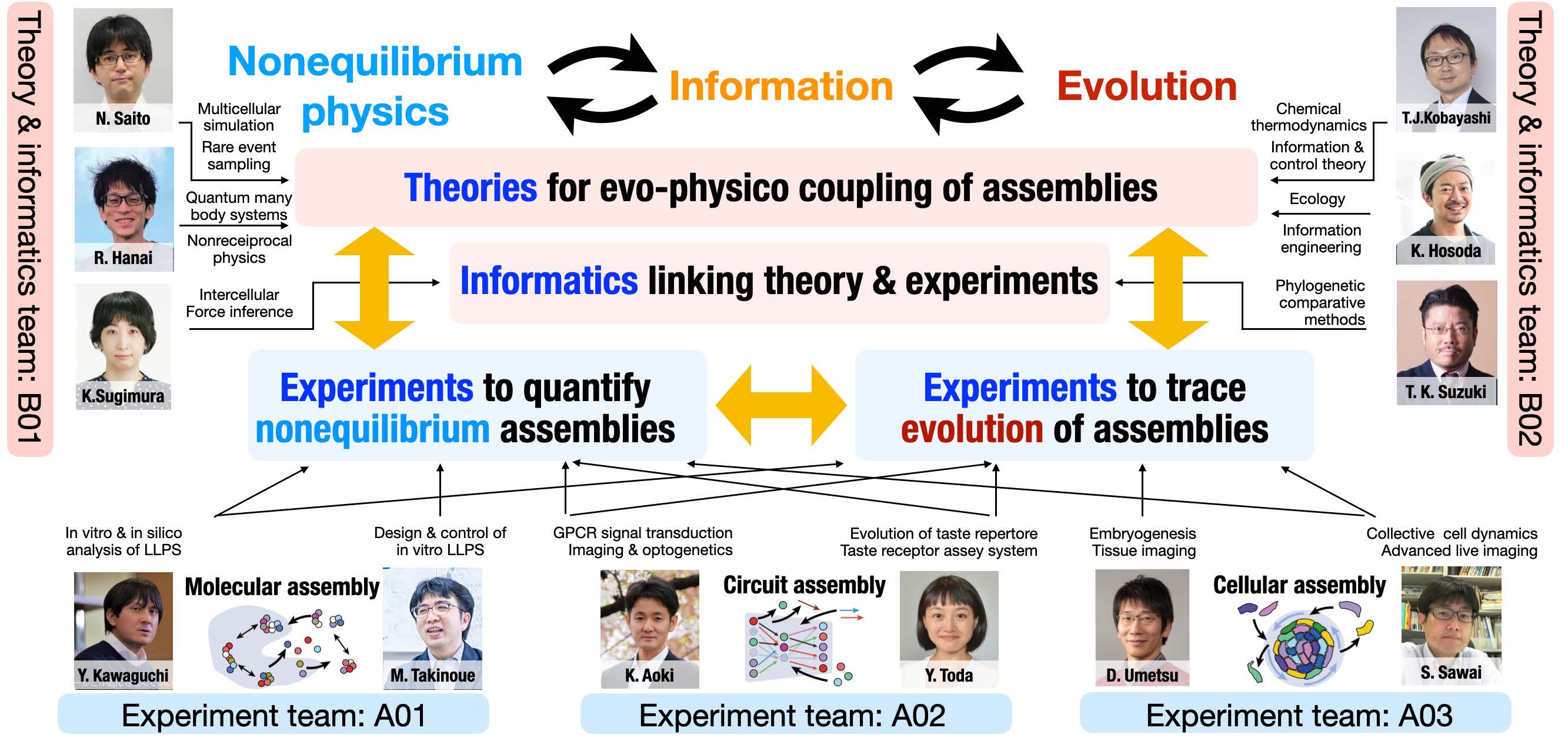
Team organization
To achieve this strategy, we organize five main projects and teams:
- Kyogo KAWAGUCHI : Chief Scientist (RIKEN), Associate Professor (University of Tokyo)
- Masahiro TAKINOUE : Professor (Institute of Science Tokyo)
- Kazuhiro AOKI : Professor (Kyoto University)
- Yasuka TODA : Associate Professor (Institute of Science Tokyo)
- Daiki UMETSU : Lecturer (Osaka University)
- Satoshi SAWAI : Professor (University of Tokyo)
- Nen SAITO : Associate Professor (Hiroshima University)
- Ryo HANAI : Associate Professor (Institute of Science Tokyo)
- Kaoru SUGIMURA : Associate Professor (University of Tokyo)
- Tetsuya J. KOBAYASHI : Professor (University of Tokyo)
- Takao K. SUZUKI : Assistant Professor (Juntendo University)
- Kazufumi HOSODA : Principal Investigator (CiNet, NICT)
Information
EPIC Office
epic.assembly.jp★gmail.com
(Please replace ★ with @ when sending.)